By leveraging human genetics to identify drug targets with a proven link to disease mechanisms, the probability of clinical trial success greatly increases [1].
Only 10% of drugs that enter phase 1 clinical trials eventually make it to market [2], primarily due to issues related to efficacy or safety that arise later on. These frequent clinical failures highlight the significant opportunity for enhancing target selection and improving clinical outcomes. Drugs that target mechanisms backed by GWAS evidence are twice as likely to receive approval [1]. Furthermore, a comprehensive review of FDA drug approvals revealed that drugs supported by human genetic data are 2 to 5 times more likely to result in successful therapies [3].
Why is human genetic data essential for improving target selection?
The development of effective drugs is often hindered by the challenge of selecting the right target. To increase the chances of success, it’s important to base target selection on a deep understanding of disease mechanisms. Genetic data can provide valuable insights into the mechanisms underlying diseases, establishing a causal link between the gene product’s function and the disease phenotype. By understanding this link, you can confidently select the right target and develop drugs that modulate it, thereby impacting the disease processes. This understanding also aids in choosing the appropriate treatment modality.
What methods can be used to validate the potential of a drug target beyond human genetics?
Apart from human genetics, other experimental data from cellular and animal systems can also inform target selection. These experiments help validate the target’s role in disease pathophysiology, further strengthening the causal link between the gene product’s function and the disease phenotype. A solid understanding of this link, leads to therapeutic interventions that address the disease’s underlying mechanisms.
Finding genetic and experimental data to support drug target selection
DISGENET is an extensive and reliable gene-disease database that contains information on the molecular mechanisms of all human diseases for a large catalog of coding and non-coding genes, as well as genomic variants. By using DISGENET, you can search for potential drug targets for any indication.
Genetic support for FDA-approved drugs 2023
In order to illustrate the information DISGENET can provide, we explored the available genetic evidence on the platform for 2023 FDA-approved drugs.
Our retrospective analysis found that DISGENET supports a connection between drug targets and therapeutic indications for a large majority of 2023 FDA-approved drugs (91%). In fact, the same analysis for 2021 showed that DISGENET provides genetic evidence for 90% of approved drugs, outperforming other publicly available platforms, which covered only 66%.
Start your 7-day free trial of DISGENET Advanced
Leveraging genetics in drug discovery: Examples from 2023 FDA-approved drugs
Targeting the protein coded by a specific gene known to cause a particular disease greatly enhanced the probability of clinical trial success [4]. DISGENET provides a comprehensive list of genes associated with a disease or indication. DISGENET also includes metrics such as the Gene Disease Association (GDA) score (ranging from 0 to 1) to prioritize those genes/proteins by disease/indication, and provide several data attributes to assist the user in understanding the role of the gene in the disease.
Skyclarys and Friedreich Ataxia
Most cases of Friedreich Ataxia (FA) are due to defects in the FXN gene that lead to depletion of FXN and impairment of antioxidant pathways in the cell. DISGENET contains 185 genes associated with Friedreich Ataxia, the highest scoring association being FXN. Notably, DISGENET ranks NFE2L2, which codes for NRF2, as the second-highest scoring associations with FA.
NRF2 is the target of Skyclarys (Omaveloxolone), the first approved treatment for this disease. Most of the evidence supporting the association of this protein and FA has been captured using text mining from articles published between 2013 and 2023.
NRF2 is involved in the regulation of the oxidative stress pathway, one of the mechanisms underlying the disease. DISGENET provides information about the role of the NFE2L2 gene and the pathogenesis of the disease.
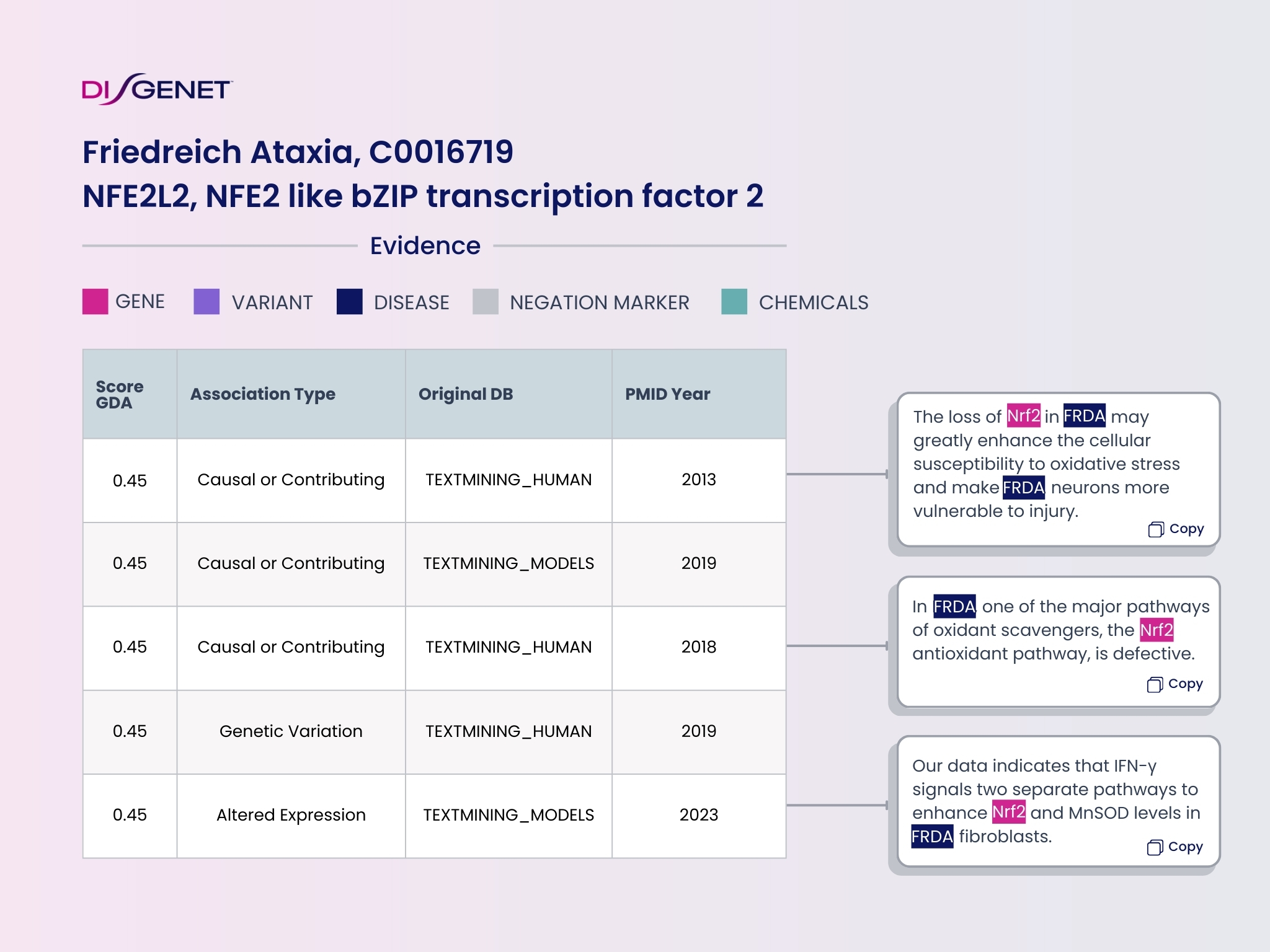
This information is also represented in the DISGENET Association Type. In this case, one of the association types for NFE2L2 and FA is “altered expression”, which is in agreement with the observation that the expression of NRF2 is decreased in skin biopsies in FA patients. NRF2 is also involved in mitochondrial homeostasis, maintenance of GSH levels and neurogenesis.
Thus, DISGENET is a key resource for providing mechanistic information on potential gene targets and therefore expediting the selection of relevant drug targets.
Veozah and Hot Flushes
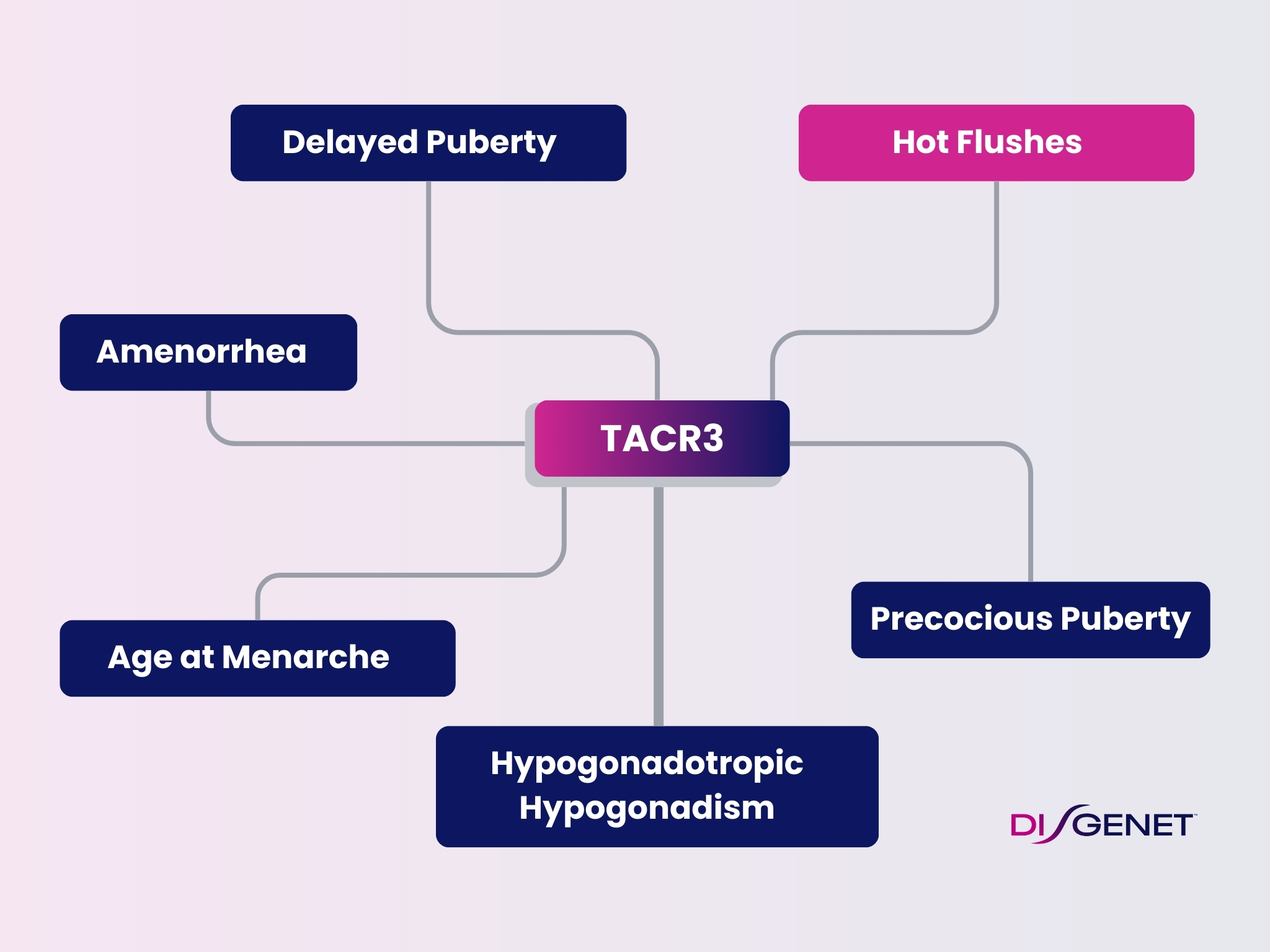
Veozah (Fezolinetant), approved for the treatment of Hot flushes, targets the Neuromedin-K receptor encoded by the TACR3 gene. DISGENET contains information on the association between the gene and the indication, with evidence dating back to 2017, including publications supporting the role of antagonists of the receptor for ameliorating hot flushes in menopausal women.
This text-mined data, identified using DISGENET’s state-of-the-art NLP tool, is not present in expert-curated databases such as UniProt or ClinGen. Variants in the gene have also been reported as associated with these symptoms. Interestingly, this gene and its variants are also associated with related traits, such as amenorrhea, age at menarche, hypogonadism, and alterations of puberty, underscoring the role of TACR3 in reproductive health.
Leveraging genetics in drug discovery using DISGENET
The above examples illustrate the valuable information that DISGENET can provide, allowing you to expedite drug target selection programs.
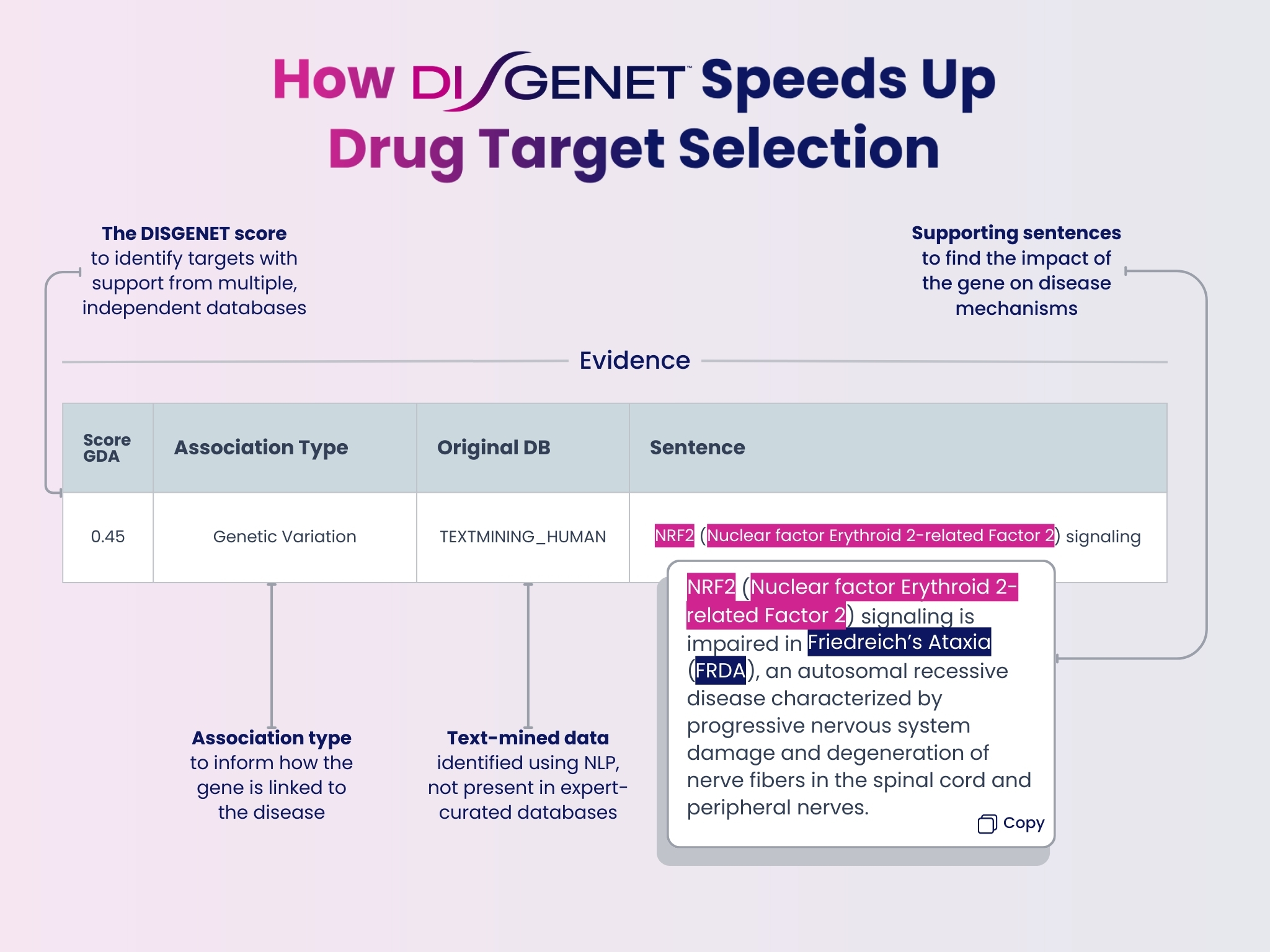
The DISGENET platform offers:
- The DISGENET GDA score to identify targets with support from multiple, independent databases.
- Text-mined data not present in expert-curated databases.
- Association type to inform how the gene is linked to the disease.
- Supporting sentences providing an easy way to find the impact of the gene on disease mechanisms.
- Comprehensive gene-disease profiles to help identify potential therapeutic targets and anticipate safety risks.
- Loss-of-function/gain-of-function annotations and variant database information to aid in drug target nomination.
Start searching DISGENET here (no account needed)
Book a Free Demo here
Contact us at info@disgenet.com
References
[1] Cook, D., Brown, D., Alexander, R., March, R., Morgan, P., Satterthwaite, G. et al. (2014) Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nature Reviews Drug Discovery, 13, 419–31. https://doi.org/10.1038/nrd4309
[2] Minikel, E.V., Painter, J.L., Dong, C.C. and Nelson, M.R. (2024) Refining the impact of genetic evidence on clinical success. Nature, 629, 624–9. https://doi.org/10.1038/s41586-024-07316-0
[3] Nelson, M.R., Johnson, T., Warren, L., Hughes, A.R., Chissoe, S.L., Xu, C.-F. et al. (2016) The genetics of drug efficacy: opportunities and challenges. Nature Reviews Genetics, 17, 197–206. https://doi.org/10.1038/nrg.2016.12
[4] King, E.A., Davis, J.W. and Degner, J.F. (2019) Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. Marchini J, editor. PLOS Genetics, 15, e1008489. https://doi.org/10.1371/journal.pgen.1008489
